Ayush's work on AnswerThis.io stems from deep research into making large language
models more effective for
scientific use. This includes developing robust customer-facing AI systems that
generate outputs with clear
citations and precise attributions, ensuring researchers can trust the
results.
His research focuses on deep analysis of research papers to extract critical
insights and applying real-time
reranking algorithms to produce coherent research summaries. These systems are
designed to highlight key points with
a deep understanding of the source material, revolutionizing the way researchers
interact with and interpret vast
bodies of academic literature.
The innovations behind AnswerThis include creating scalable AI systems capable of
handling complex queries,
integrating real-time feedback loops, and enhancing user experience by streamlining
how insights are presented.
These efforts are advancing how AI can support the academic community.
Ayush's research with Dr. Catherine Finegan-Dollak at the University of Richmond
focuses on utilizing grounded NLP to translate human instructions into
computer-executable commands. This research aims to bridge the gap between human
communication and machine understanding by allowing for more efficient and effective
interactions between the two.
The research is focused on developing natural language understanding systems that
can accurately interpret human instructions and execute them in a simulated
environment, such as a video game. This involves using techniques such as language
grounding, semantic parsing, and reinforcement learning. The goal is to create
systems that can understand and act on human instructions in a way that is more
natural and intuitive for humans.
In summary, Ayush's research on grounded NLP aims to develop natural language
understanding systems that can accurately interpret human instructions and execute
them in a simulated environment, thus allowing for more efficient and effective
interactions between humans and machines.

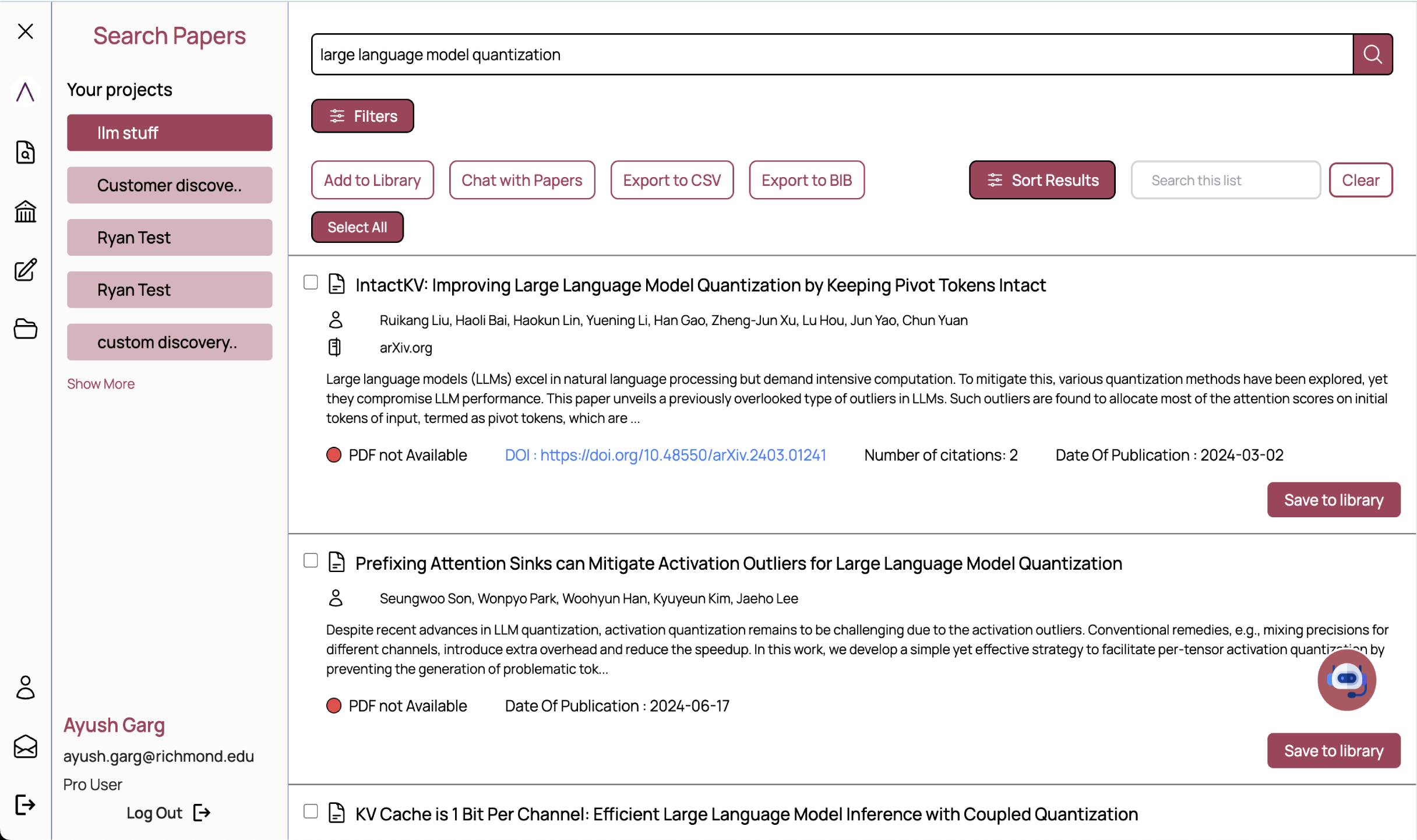
pygetpapers
has been developed to allow searching of the scientific literature in
repositories
with a range of textual queries and metadata. It downloads content using APIs in
an automated
fashion and is designed to be extensible to the growing number of Open Access
repositories.
This JOSS
article further elaborates the design of the tool.
An increasing amount of research, particularly in medicine and applied science,
is now based on meta-analysis and sytematic review of the existing literature
(example). In such reviews scientists
frequently download thousands of articles and analyse them by Natural Language
Processing (NLP) through Text and Data Mining (TDM) or Content Mining (ref). A
common approach is to search bibliographic resources with keywords, download the
hits, scan then manually and reject papers that do not fit the criteria for the
meta-analysis. The typical text-based searches on sites are broad, with many
false positives and often only based on abstracts. We know of cases where
systematic reviewers downloaded 30,000 articles and eventually used 30.
Retrieval is often done by crawling / scraping sites, such as journals but is
easier and faster when these articles are in Open Access repositories such as
arXiv, Europe/PMC biorxiv, medrxiv. But each repository has its own API and
functionality, which makes it hard for individuals to (a) access (b) set flags
(c) use generic queries.
In 2015 we reviewed tools for scraping websites and decided that none met our
needs and so developed getpapers, with the key advance of integrating a query
submission with bulk fulltext-download of all the hits. getpapers was written in
NodeJs and has now been completely rewritten in Python3 (pygetpapers) for easier
distribution and integration. Typical use of getpapers is shown in a recent paper where the authors
"analyzed key term frequency within 20,000 representative [Antimcrobial
Resistance] articles".

Unsupervised entity extraction from sections of papers that have defined
boilerplates. Examples of such sections include - Ethics Statements, Funders,
Acknowledgments, and so on.
Primary Purpose:
Extracting Ethics Committees and other entities related to Ethics Statements
from papers
Curating the extracted entities to public databases like Wikidata
Building a feedback loop where we go from unsupervised entity extraction to
curating the extracted information in public repositories to then, supervised
entity extraction.
Subsidary Purpose(s):
The use case can go beyond Ethics Statements. docanalysis is a general package
that can extract relevant entities from the section of your interest.
Sections like Acknowledgements, Data Availability Statements, etc., all have a
fairly generic sentence structure. All you have to do is create an ami
dictionary that contains boilerplates of the section of your interest. You can,
then, use docanalysis to extract entities. Check this section which outlines steps for
creating custom dictionaries. In case of acknowledgements or funding, you might
be interested in the players involved. Or you might have a use-case which we
might have never thought of!



 Python
Python
 Git
Git
 HTML5
HTML5
 CSS3
CSS3
 JavaScript
JavaScript
 React
React
 Java
Java